miRNome analysis using Affymetrix microarrays:
Bioalternatives offers different solutions for microRNA analysis. The analysis of the whole microRNA expression profile (miRNome), using a DNA microarray (miRNA 4.1), will detect the molecular signatures and identify microRNAs of interest.
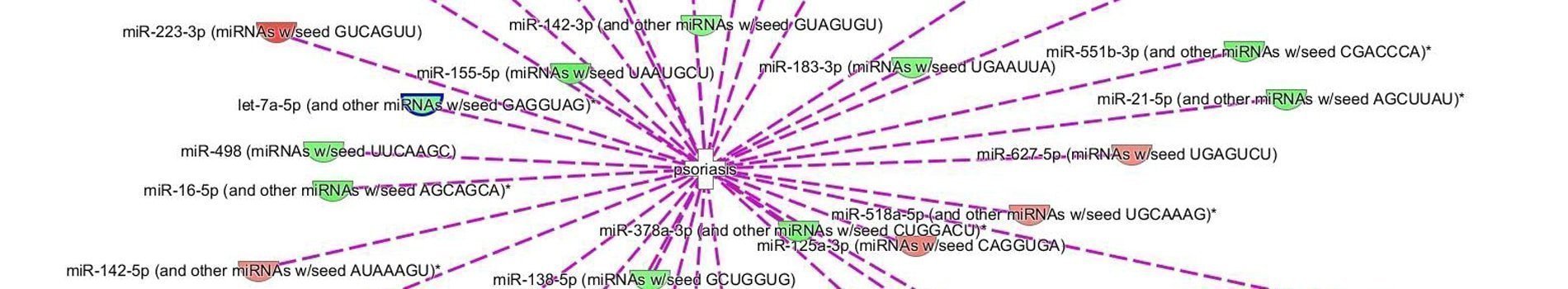
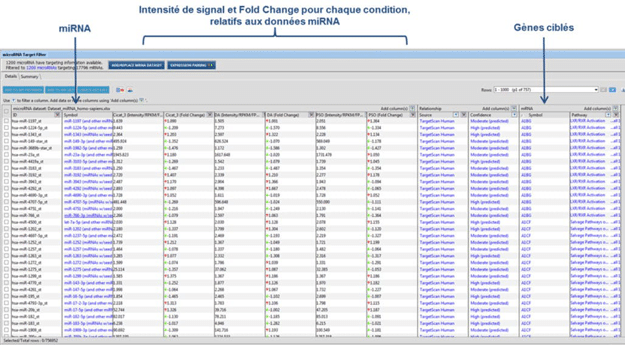
miRNome analysis from Affymetrix miRNA4.1 microarray will allow the identification of significantly modulated genes with fold change calculation and statistical tests (t-test performed with replicates), which will detect the characteristic molecular signature of the sample (figure 1). A second step of analysis will identify the target genes of significantly modulated miRNAs, using Qiagen IPA® software (figure 2).
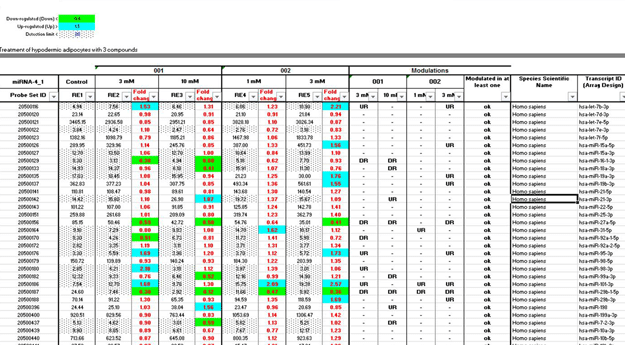
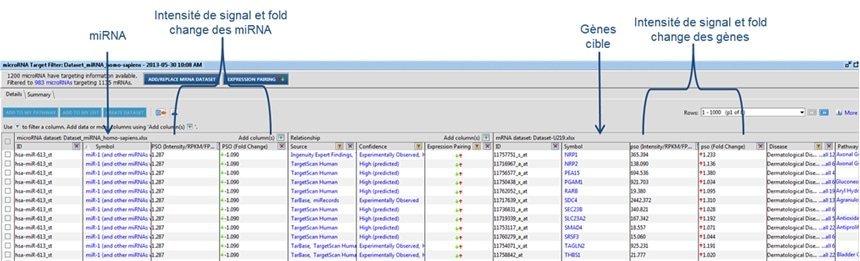
Figure 1: Table of result analysis
The matrix calculation in this table allows the calculation of fold change values and the detection of significantly modulated miRNAs.
Figure 2 : Identification of miRNA targeted genes using Qiagen IPA® software
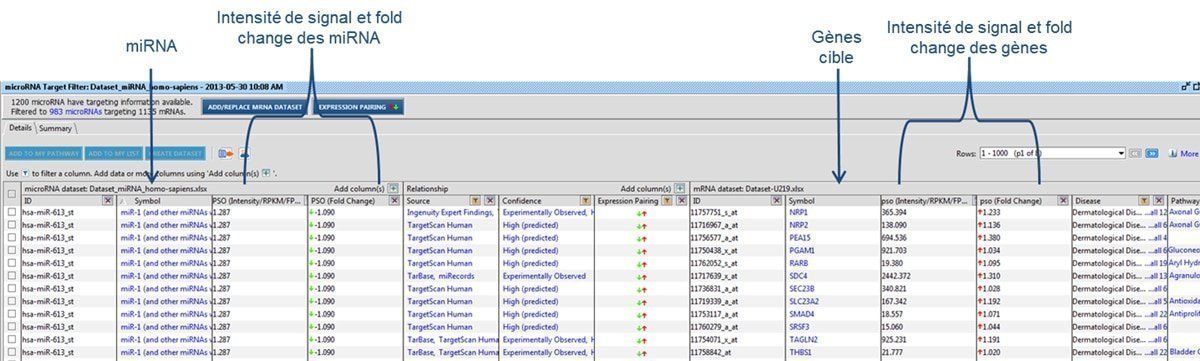
For studies of the regulation mechanism using miRNAs, miRNome analysis may be performed along with a transcriptome analysis, by using a U219 microarray (human microarray) on the same samples. The hybridization of the same sample on two types of microarrays will allow the identification of the significantly modulated genes that are potentially regulated by miRNAs. Correspondence of U219 and miRNA4-1 microarray data is determined by using Qiagen IPA® software (Figure 3).
Figure 3 : Correspondence of U219 and miRNA4-1 microarray data, using Qiagen IPA® software
MiRNA expression analysis by RT-qPCR:
RT-qPCR experiments analyse and compare the expression level of targeted genes in given conditions. The same method can be used for miRNA analysis.
Bioalternatives can perform the extraction and the analysis of targeted miRNA expression by RT-qPCR.
Applications:
- Biomarker validation
- Diagnosis and prognosis
- Research of target-genes
- Confirmation of analysis by microarrays
Characteristics:
- High sensitivity
- High specificity
- Speed
Principle:
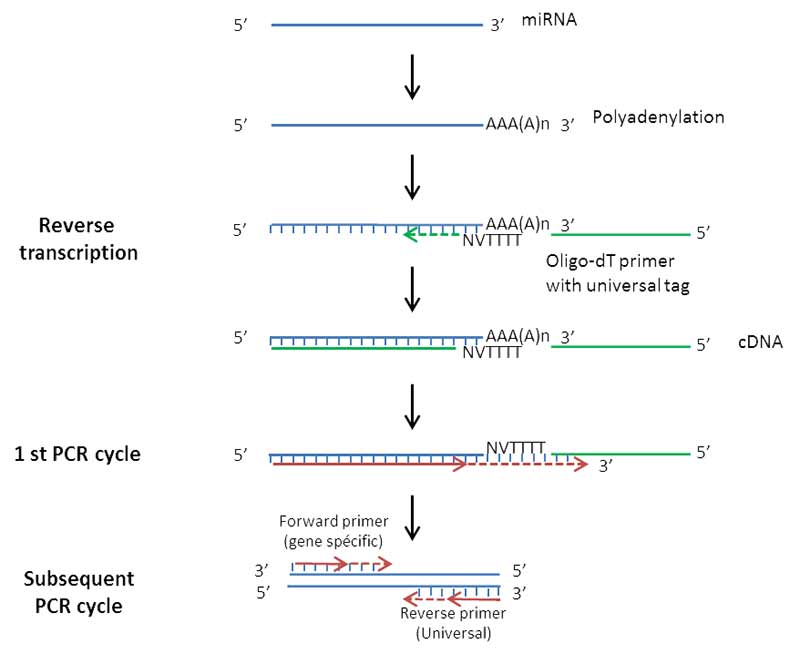
- Extraction of total RNA and purification of miRNA, using spin columns
- Polyadenylation of miRNA
- Reverse transcription using an Oligo-dT primer
- PCR amplification using a specific primer and a universal primer
- Measurement of the incorporation of fluorescent molecules
Figure 1: Principle of miRNA analysis by RT-qPCR
Analysis and result interpretation:
Result analysis is based on a continuous measurement of fluorescent molecule incorporation (SYBR Green) during the amplification process of the PCR reaction.
These measures are transcribed into numerical values (number of cycles) for each gene. These values are then forwarded and processed under Microsoft Excel in order to calculate the relative expression level compared to a control condition (Figure 2).
In Bioalternatives, we have the necessary bioinformatics resources and know-how to help you go further by identifying the target-genes of miRNA of interest.
Figure 2: Identification of target-genes