The skin microbiota plays a pivotal role in maintaining skin health and balance. This dynamic ecosystem of bacteria, fungi and viruses lives in symbiosis with skin cells, contributing to barrier function, immune defense, and overall homeostasis.
Disruptions to this delicate equilibrium – whether due to pollution, harsh cosmetics, aging, or lifestyle – can lead to dysbiosis. This microbial imbalance has been associated with a range of skin conditions, including dryness, sensitivity, acne, and atopic dermatitis.
In recent years, the concept of “microbiome-friendly” skincare has emerged as a major trend in dermo-cosmetics. It reflects a growing consumer awareness of the skin’s invisible allies and a desire for gentler, more holistic beauty routines.
At QIMA Life Sciences, we empower our clients by providing innovative models and assays tailored to the study of host – microbiome interactions. Our expertise in skin biology, molecular analysis, and imaging technologies makes us the ideal partner for microbiota-centred product innovation.
We Support You at Every Stage of the Innovation Cycle to Substantiate Efficacy Claims.
We Support You Through Every Stage of Product Development
Skin Microbiota: Available Models & Assays
Thanks to its expertise in cellular and tissue engineering, QIMA Life Sciences has developed in vitro or ex vivo study models that are suitable for analyzing the interactions between microbiota and skin:
- Isolated bacterial culture (Staphylococcus aureus, Staphylococcus epidermidis, Cutibacterium acnes, Corynebacterium xerosis …)
- Co-culture:
- bacteria on 2D skin models (keratinocytes, fibroblasts, sebocytes, etc.)
- bacteria on 3D skin models (reconstructed epidermis, etc.)
- bacteria on human skin explants (human full thickness skin)
- Infection experiments with the HSV-1 virus ( Herpes simplex virus type I)
Both the impact of compounds or formulations on the development of bacteria and the influence of bacteria on cutaneous response can be assessed by these models.
QIMA Life Sciences offers a panel of solutions for the evaluation of:
- Bacterial viability
- Quantification of bacterial colony (CFU)
- Determination of bacterial load (qPCR (Taqman))
- Bacterial adhesion on 3D models (reconstucted epidermis) – quantification of colony, qPCR, radioactivity, SEM
- Biological response of 2D or 3D models (differentiation, inflammation and immune response)
- Investigation of 16S rRNA and certain bacterial strains in situ (In situ hybridization)
-
Bacterial community analysis (Bacterial 16S rRNA sequencing)
-
Fungal/microbial community analysis (ITS sequencing/Shotgun sequencing)
-
Prevention of hair follicle/skin microbial overgrowth (contamination)
-
Expression of antimicrobial peptides
-
Manipulation of microbiome using antibiotics
-
Agar diffusion assay of selected bacterial and fungal strains using culture medium
In addition, laser capture microdissection allow us to perform analysis in selected skin explants compartments using tissue sections.
QIMA Life Sciences provides a wide range of microorganisms tested by our laboratory and available for in vitro testing:
BACTERIA
GRAM (+) :
- Staphylococcus aureus
- Staphylococcus epidermidis
- Cutibacterium acnes
- Corynebacterium xerosis
- Streptococcus pyogenes
- Mix of several strains (upon request)
- Other strains upon request
GRAM (-) :
- Pseudomonas aeruginosa
- Other strains upon request
VIRUSES
- Herpes simplex virus type 1 (HSV-1)
- Human Rhinovirus 16 (HRV-16)
YEASTS

Coming soon
Skin microbiota analysis
Our company has developed ready-to-use non-invasive collection kits to analyze the lipids and biomarkers of the skin surface from your samples or from those of your clinical center.
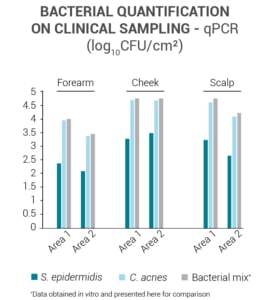
The qualitative and quantitative analysis of the bacteria present on the surface of the skin are carried out on samples with the SW Kit. They are performed using the following techniques:
- Traditional microbiology on agar
- Targeted qPCR
- Non-targeted metagenomic analysis
In addition, the analysis of sebaceous lipids (triglycerides and fatty acids) gives information on the lipase activities of bacteria and yeasts such as C. acnes and M. furfur, involved in the development of acne and dandruff respectively.
These evaluations help support your claims about the efficacy of “friendly” microbiota products, bacteriostats, antimicrobials, prebiotics, anti-acne products, anti-dandruff products, etc.
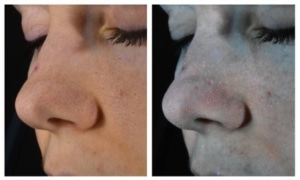
Data Mapping & Clinical Imaging
Measurement of porphyrin fluorescence in UV imaging
Acne-prone skin can present a high fluorescence rate under UV, due to the metabolism of the bacteria specific to acne. The measurement of porphyrin fluorescence intensity is therefore a marker of the evolution of the inflammatory and bacterial skin condition.
Measurement of skin fluorescence on image acquired with ColorFace ®
At QIMA Life Sciences, we specialize in providing comprehensive solutions for studying skin aging and longevity through validated models at both preclinical and clinical levels.
Our dedication to scientific innovation drives us to develop cutting-edge models and approaches, staying aligned with the latest advancements in longevity research.
Interested in Learning More?
Explore Other Related Topics
HAIR FOLLICLE MICROBIOME






